I’m trying to build a better personal repository of biology datasets on a remote server that has plenty of storage. One idea I had is to construct a big list of models and then systematically test each model against each of the datasets as I add them, and see which one works best . I was inspired by someone I spoke to at a dinner regarding fully automated science, and so I decided to have a go.
So today, at the London Independent Science Society chill coffee session, I rented a Hetzner server (around 30GBP a month) to download all of the data from the Biotime (Dornelas, M., Antão, L.H., Moyes, F., Bates, A.E., Magurran, A.E., et al. (2023))package, which is a 1GB collection of ecological time-series aggregated across different geographies. My main motivation was to find some datasets that were amenable to dynamic mode decomposition, or some form generalised Lotka-Volterra modelling. I’ve written quite a bit on toy Lotka-Volterra models for ecology, and I thought that this would be a good place to test out those models on some real world data.
Out of the thousands of datasets in the package, I tried to find some that were large enough in quality to do some real modelling work on. But when I dug into it, I struggled to find a dataset that suited my purposes. Unfortunately, a lot of the datasets were just observatory data, and it was a bit tricky finding a set that looked at a single ecosystem, with persistent, stable species measured at regular time intervals.
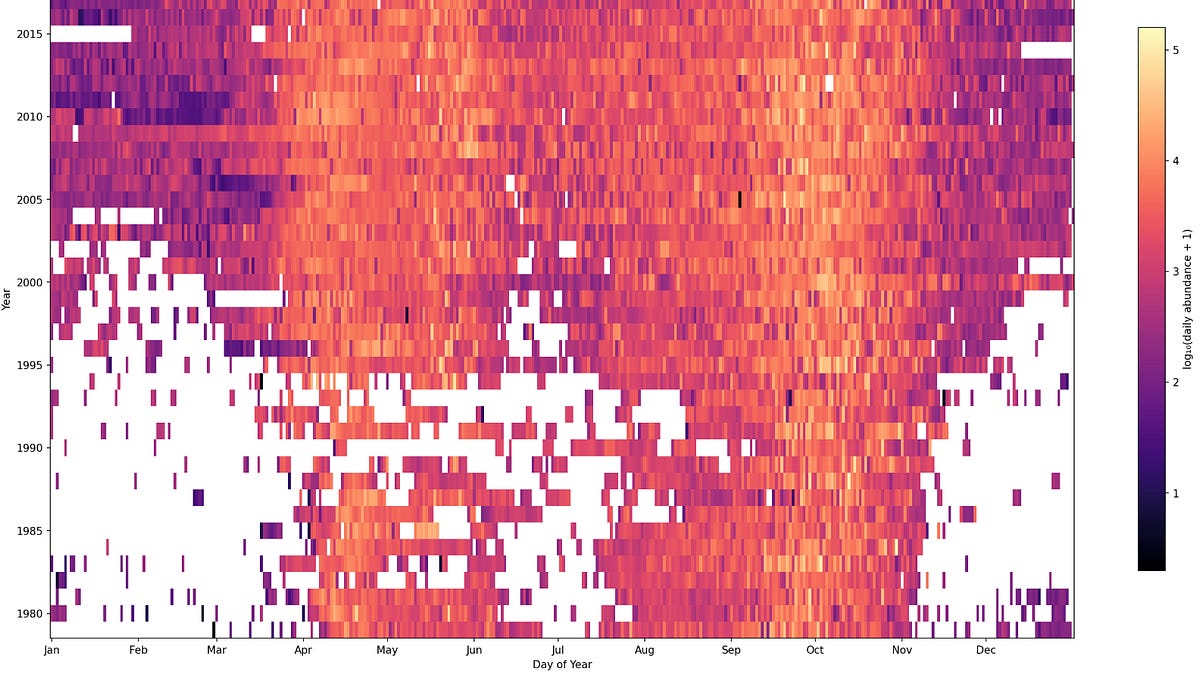
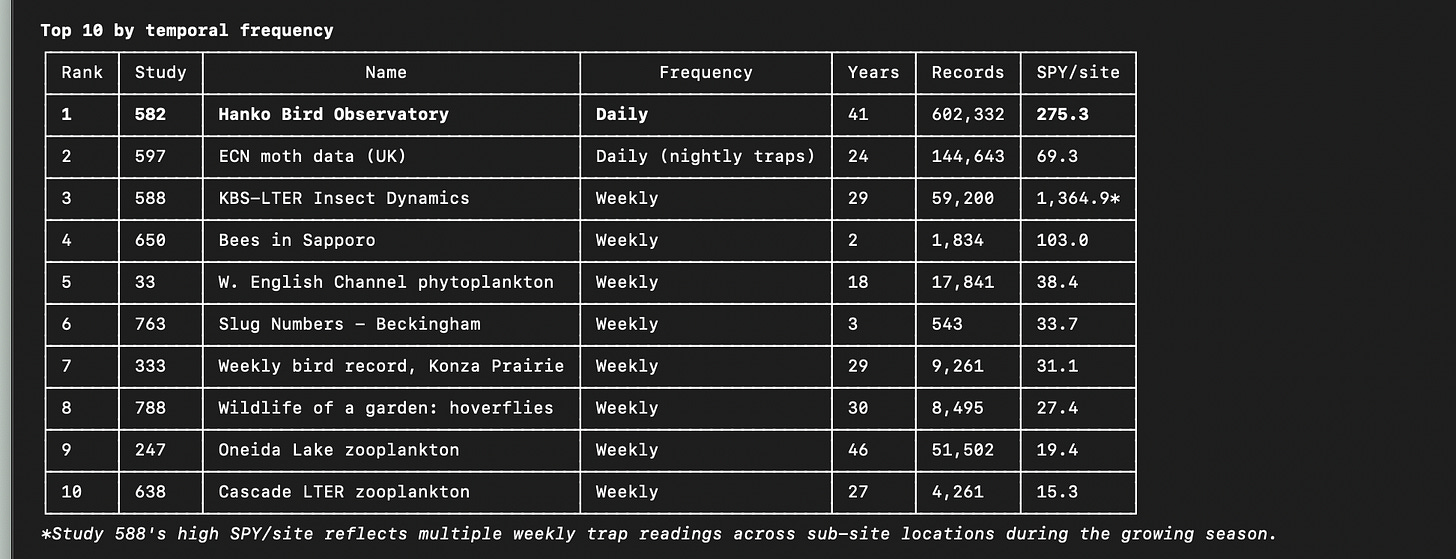
Here is a summary of some of the interesting datasets I found in the whole collection, where I intentionally looked for datasets with the highest frequency of observation.
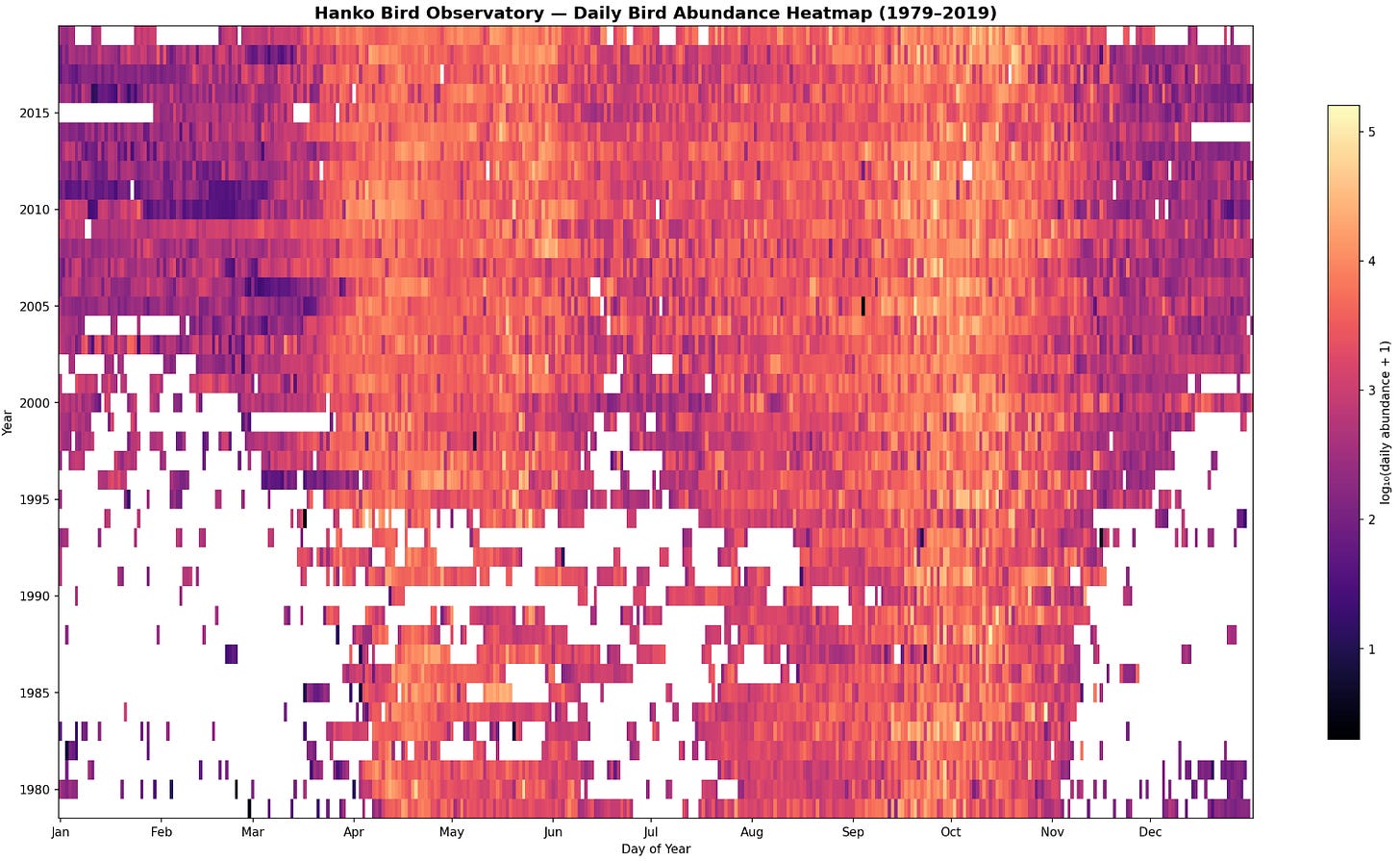
An example, the presence of birds’ migratory patterns throughout the seasons as observed from the Hanko bird observatory in Finland:
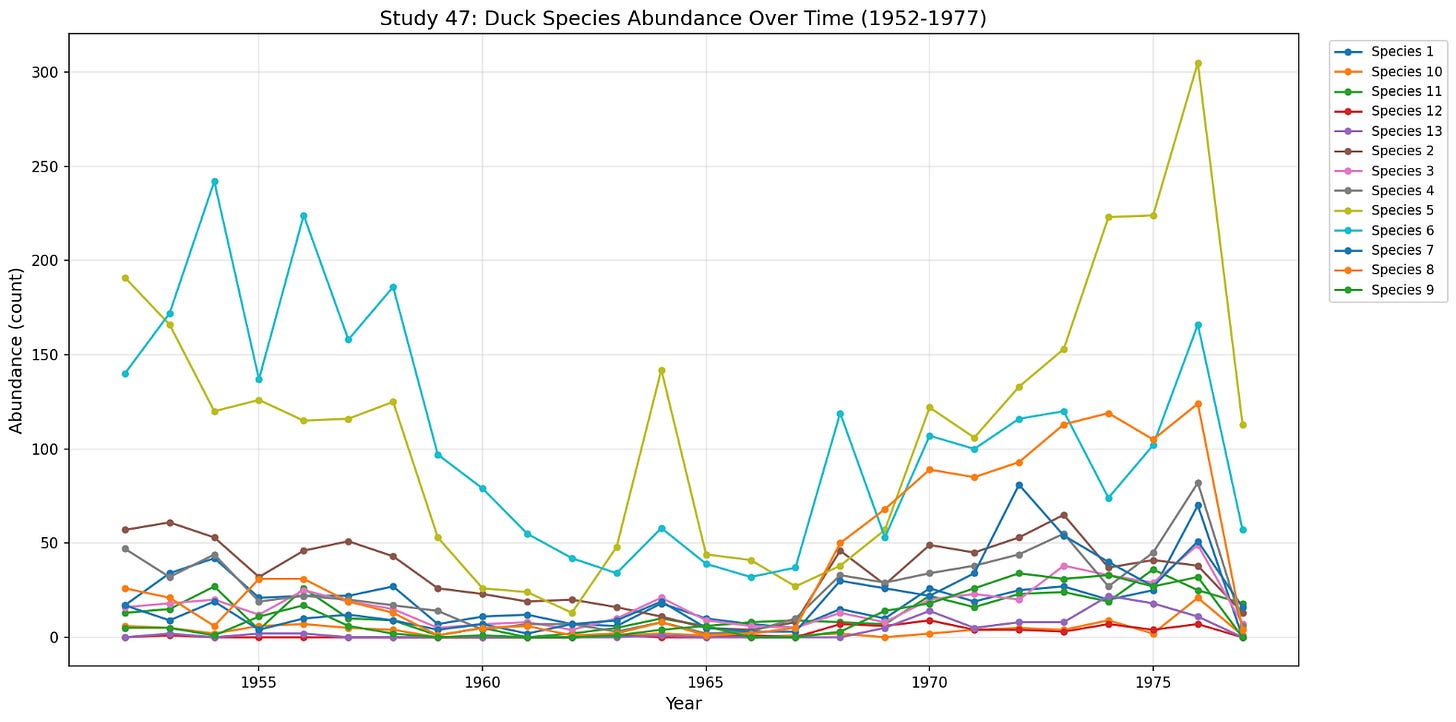
The most compelling set of data was some ecological data I found on mammals in Ontario from trapping data, and a duck dataset in Lousana and Redvers in Canada by Vickery, W.L. & Nudds, T.D. (1984). Unfortunately these again didn’t seem granular enough to do any meaningful time series analysis. Because of labour constraints, the sampling happens on the order of every year, and so even long studies that last for 30 years only have 30 samples per species, which is hardly enough to do any interesting. If you have even more than ten species but only 30 time samples, then the system is probably going to be overdetermined.
So for now, I think to do any meaningful ecology work, one needs to find a dataset with significantly more frequent sampling relative to the number of species being measured. I get that this is a hard ask, but it means that the theoretical ecology part of my work remains theoretical for now. I’ll keep looking though, since something interesting must be within this fairly large (1GB) dataset!
In other news, I started reading an old edition of Dawkins’ ‘The Selfish Gene’ today. Enjoying it so far but the paradigm of the gene encoding all information seems outdated – just read Phillip Ball’s ‘How Life Works’ to convince yourself of this. It’s also another reason for why I think dating someone on the hope of somehow passing down ‘good genes’ from them is pretty stupid for now.